Difference between revisions of "Genotyping Order Entry Help"
(→Step 4 of 10: Sample Annotation) |
(→Step 4 of 10: Sample Annotation) |
||
| Line 111: | Line 111: | ||
=='''Step 4 of 10: Sample Annotation'''== | =='''Step 4 of 10: Sample Annotation'''== | ||
| − | [[File:geno_order_entry_11. | + | [[File:geno_order_entry_11.png]] |
# These fields will be auto filled with the data provided in the Sample Input File. If any of your annotations were added into the Sample Input File, they will also be auto-filled here. If you did not provide a file, or if any information is missing, enter it now. | # These fields will be auto filled with the data provided in the Sample Input File. If any of your annotations were added into the Sample Input File, they will also be auto-filled here. If you did not provide a file, or if any information is missing, enter it now. | ||
Revision as of 17:58, 27 July 2016
Contents
- 1 Start a New Batch
- 2 Order Services - Order Type
- 3 Step 1 of 10: SNP Submission
- 4 Step 2 of 10: Batch Definition
- 5 Step 3 of 10: Sample Description
- 6 Step 4 of 10: Sample Annotation
- 7 Step 5 of 10: Sample Containers
- 8 Step 6 of 10: Select Services
- 9 Step 7 of 10: Billing Information
- 10 Step 8 of 10: SNP Selection
- 11 Step 9 of 10: Review Order
- 12 Step 10 of 10: Order Confirmation
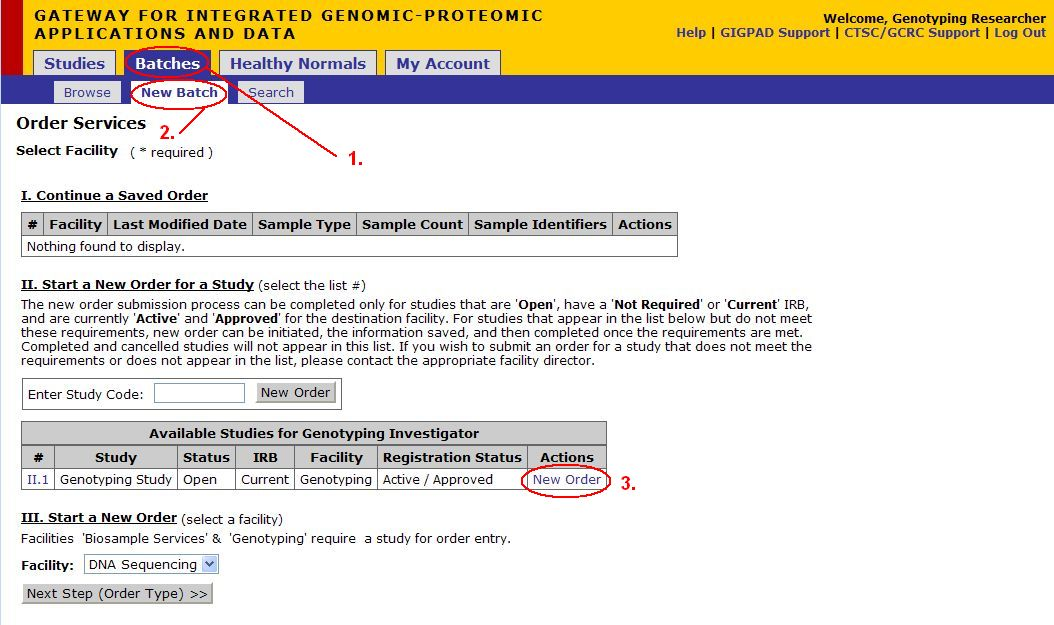
Start a New Batch
 '*Note: Genotyping Orders can be created only if study exists
'*Note: Genotyping Orders can be created only if study exists
- Click on Batches
- Click on New Batch
- Click New Order underneath the Actions column for the appropriate Study
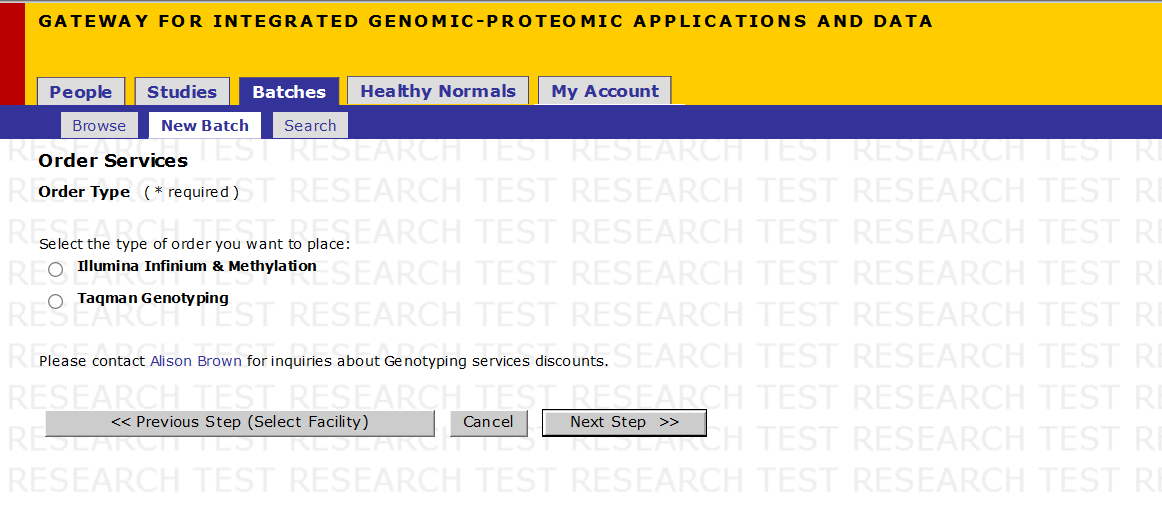
Order Services - Order Type
- Select the Order Type to be placed with the help of the radio buttons for the various platforms.
- Click on Next Step
For Ordering Services on Sequenom Platform, follow the steps below on SNP Submission. For Illumina Platform or Other Services skip to Batch Definition.
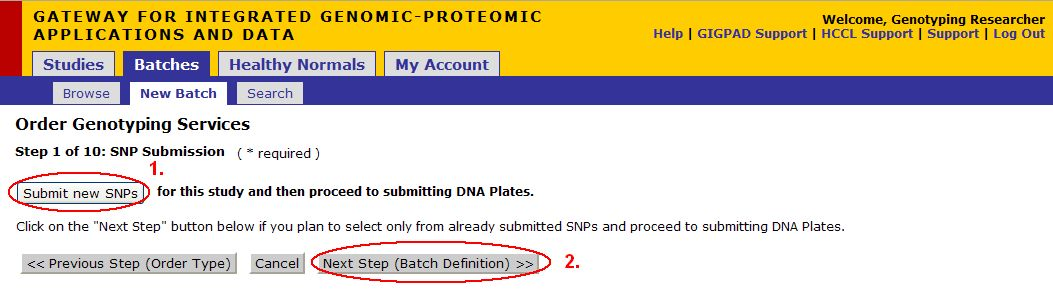
Step 1 of 10: SNP Submission
If you need to submit new SNPs for this study then:
1. Click on the Submit new SNPs button
If you plan to select only from already submitted SNPs then:
2. Click on Next Step (Batch Definition) and skip to the Batch Definition step in this documentation
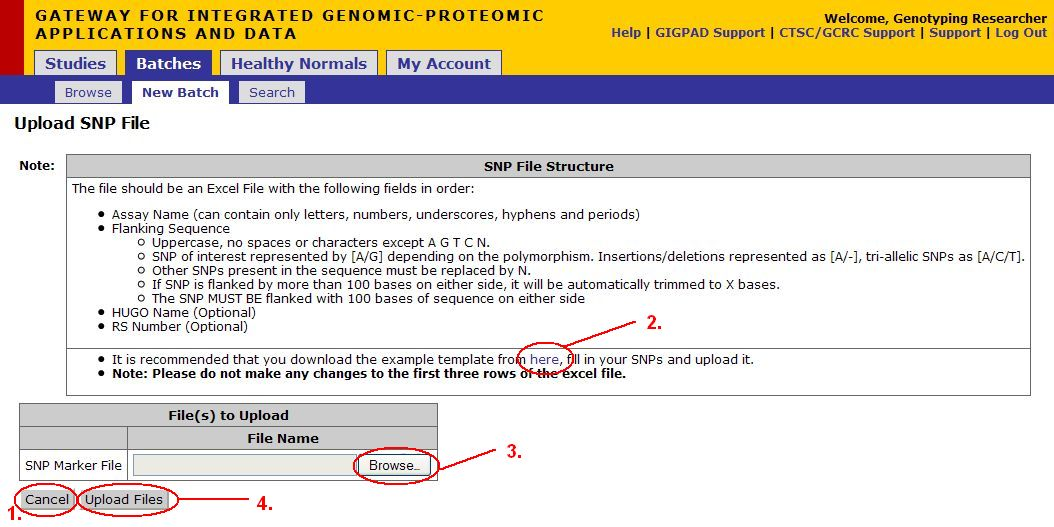
Upload SNP File
- Click on Cancel button to cancel the current operation at any time and to go back to the previous screen.
- Click on the here button (see image above) to download the template file (see next page for a more detailed description of this file)
- Once you have filled out the SNPs file, Click Browse, a dialog should appear, navigate to the location where the excel sheet you just created is stored. Select the file.
- Click Upload Files
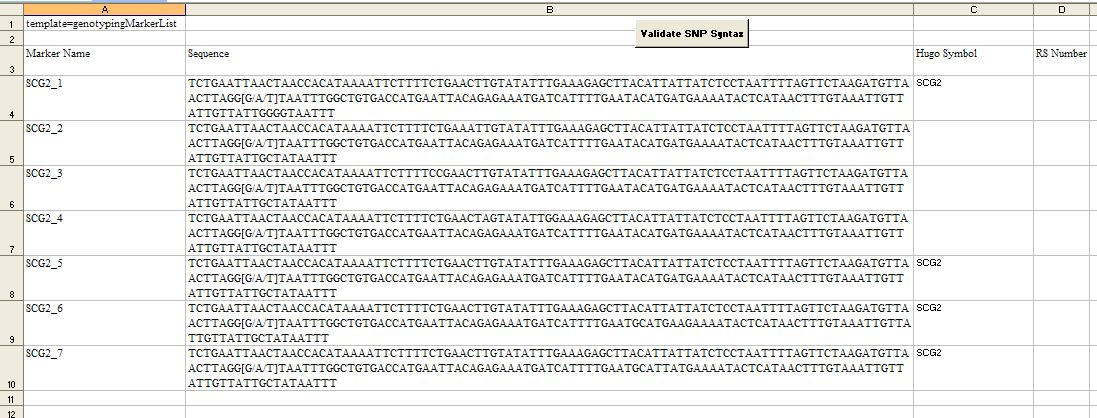
Sample SNPs File Template
- Note: Validation is also run when the file is uploaded. If the Marker Names or Sequences are incorrect, you will not be able to upload the file.
- Do not change/delete the first 3 rows in this template
- Fill in the Marker Names. The names must be EXACTLY the names of the sequences provided for assay design
- Fill in the Sequences
- Fill out any Hugo Symbols and RS Numbers (Optional)
- When you have finished entering your data, click on the Validate SNP Syntax. This will validate the following:
- Marker Name must be unique and can contain only letters, numbers, underscores, hyphens and periods with a maximum of 50 characters
- Sequences must be unique and can have only A, G, T, C and N's and the SNP of interest should be represented as [A/G] or [-/T] or [C/-] or [A/T/C]
- SNP should be flanked by at least 100 bases on either side (If the flank contains more than 100 bases, it will be trimmed down to 100 bases)
- Once validation is complete, save the template as an Excel (xls) file.
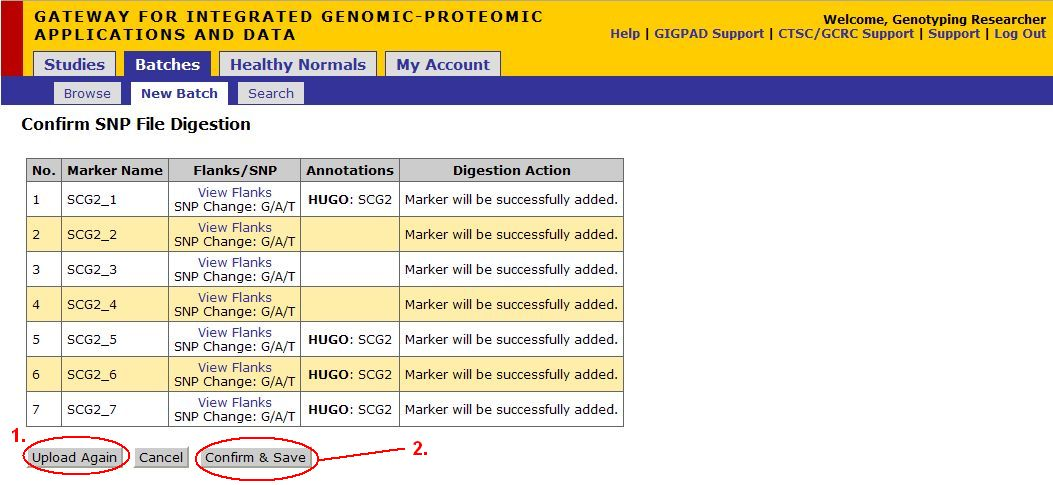
Confirm SNP File Digestion
*Note: If markers with same name are uploaded, then they would have the name appended with date. (MarkerName_mmddyy).
- To upload another SNP file, click Upload Again
- Once you have finished uploading your files. Click on Confirm and Save
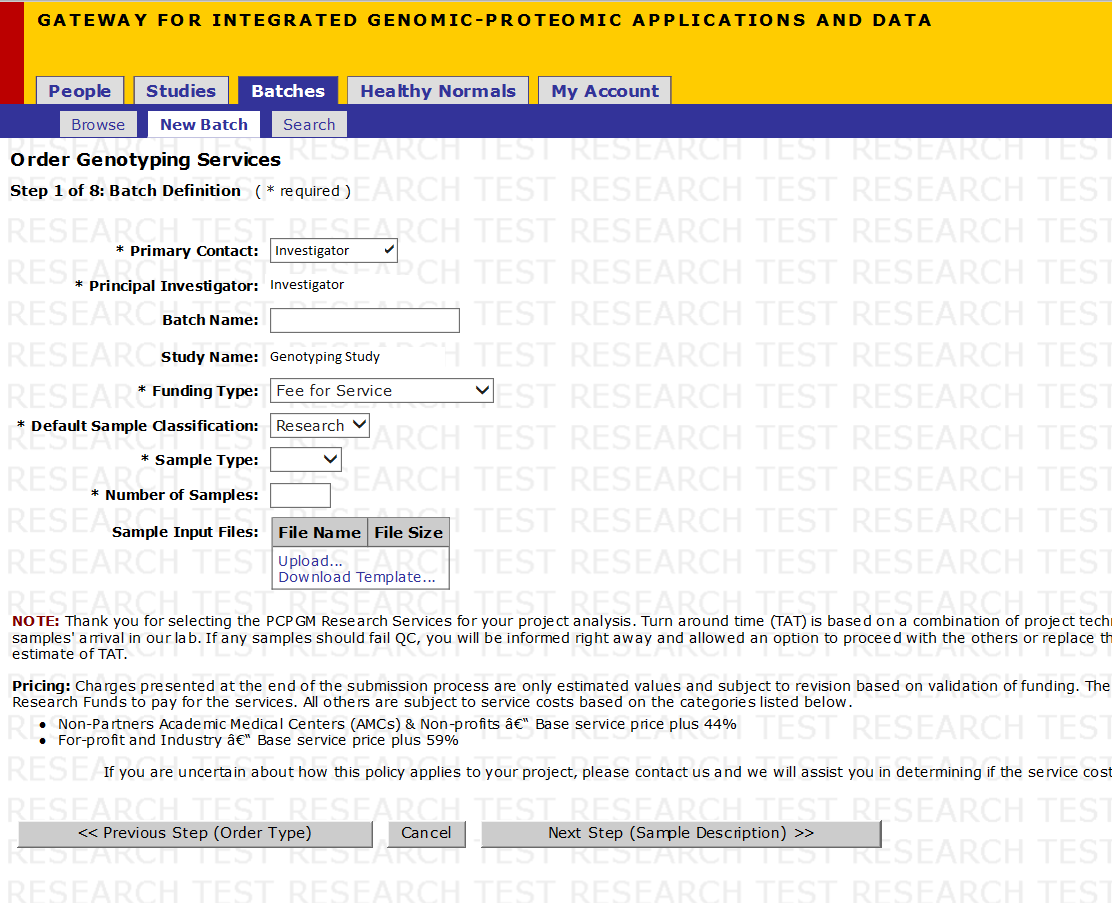
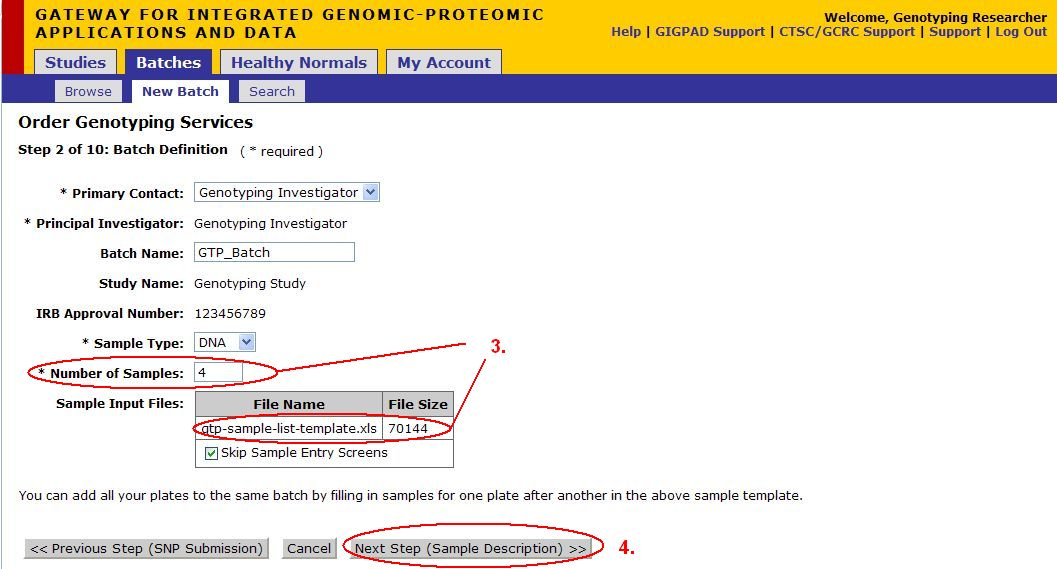
Step 2 of 10: Batch Definition
- Select Primary Contact from the drop down menu
- Select Sample Type from the drop down menu
- For a small number of samples, you can skip to Step 6 (Click on Next (Sample Description)) if you would like to manually enter your samples.
- Select Download Template... to download the Excel file that contains the template that needs to be filled out in order to upload your Sample Input File. See the next page for details on filling out this form.
- Once the form in completed, select Upload... to upload the form. See the Upload File page below for more details on the Upload screen.
- After the form has been uploaded, click on Next Step (Sample Description)
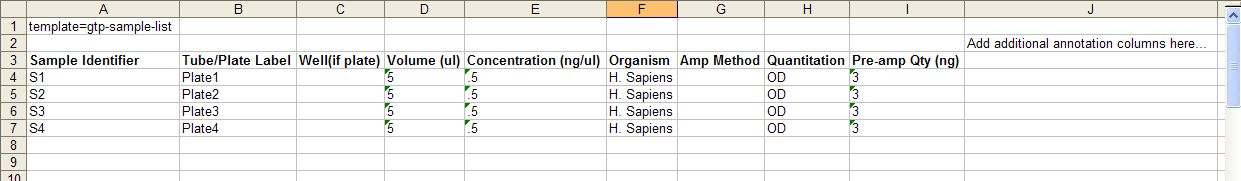
Sample Input File Template
- Sample Identifier - Sample identifier should only contain numbers, letters, dashes, underscores, parentheses, periods, and spaces
- Tube/Plate Label - Should be the same for all samples being submitted on the same plate.
Please use more complex names than “Plate1”, for example “Initials_Plate#_date”, (eg “AB_Plate1_042910”) , as we receive do receive many “Plate1”s If samples are in tubes, each tube label must be unique. Note, for Sequonom orders, the samples must be submitted on plates.
- Well – The column is required if you are submitting a plate.
- Amp method - If you are submitting genomic DNA then write "none" into this field, if you are submitting WGA DNA then please provide the method of WGA.
- Quantitation - e.g. OD, nanodrop, picogreen, gel estimation
- Pre-amp Qty (ng) - If you are submitting genomic DNA then enter “0" in this field. If you are submitting WGA DNA then please enter the quantity of DNA used in the WGA reaction.
- If you have duplicate Sample Identifiers on a Plate, then the Sample count and Well count will be different. For example, if you enter NTP as a Sample Identifier for 3 different Wells on Plate1 and Plate1 is a 96 Well Plate, then you will have a Sample Count of 94 and a Well Count of 96.
- If you wish to add annotation fields, add each annotation field as its own column, starting after the Pre-amp Qty (ng) column. Adding these into the excel file will speed up the Order Entry process, as these will be auto-filled during Step 4.
- All Other fields are optional but it is recommended that you take the time to fill out these fields now so that they are pre-filled during the following steps.
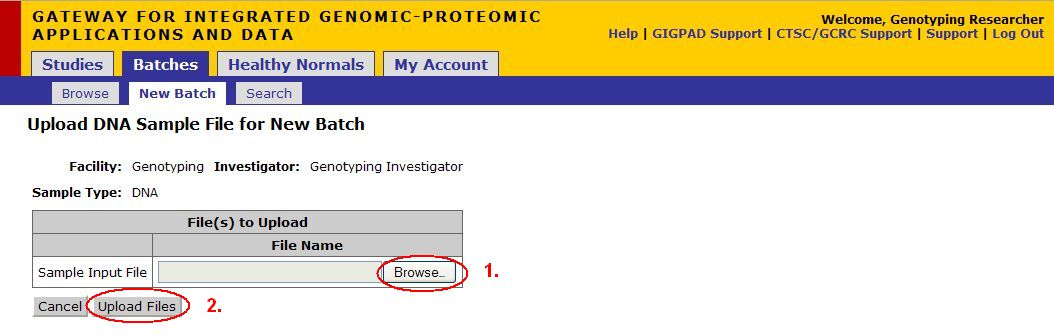
Upload File
- Click Browse, a dialog should appear, navigate to the location where the excel sheet you just created is stored. Select the file.
- Click Upload Files
- Once the upload is complete, the number of samples should appear in the Number of Samples text box and the file name should be in the Sample Input Files box
- Click on Next Step (Sample Description)
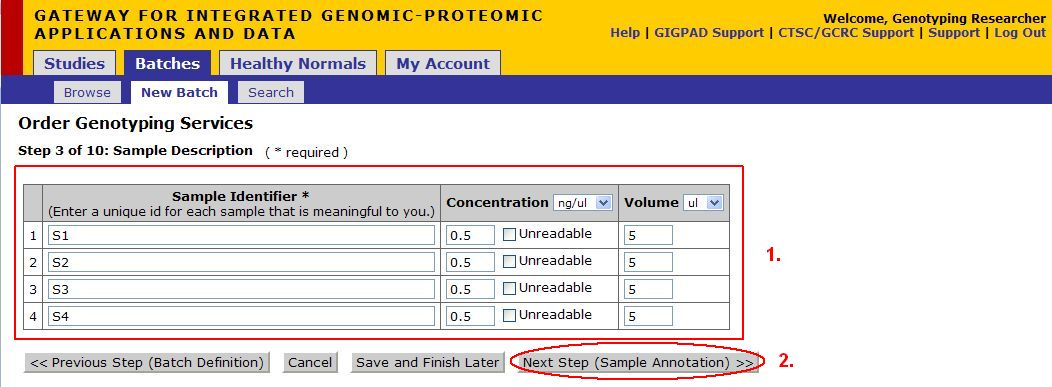
Step 3 of 10: Sample Description
- These fields will be auto filled with the data provided in the Sample Input File. If you did not provide a file, or if any information is missing, enter it now.
- Once you are satisfied with the information, click Next Step (Sample Annotation)
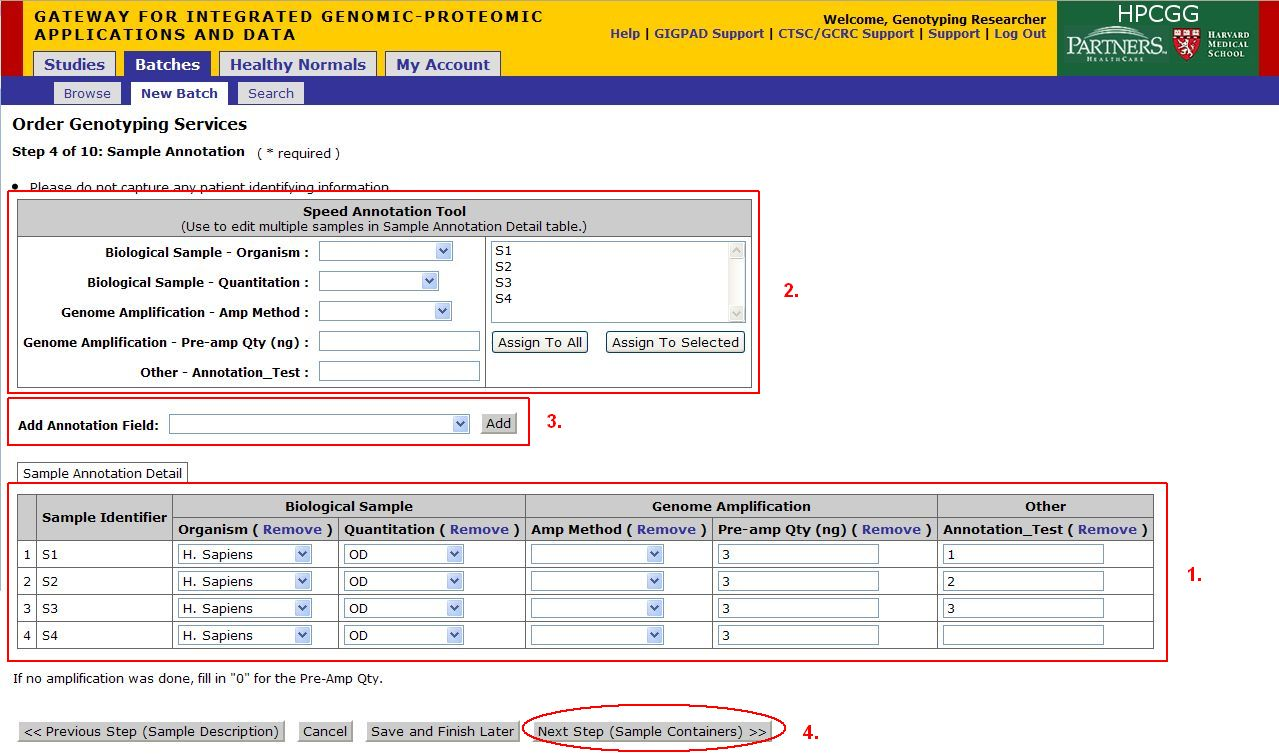
Step 4 of 10: Sample Annotation
- These fields will be auto filled with the data provided in the Sample Input File. If any of your annotations were added into the Sample Input File, they will also be auto-filled here. If you did not provide a file, or if any information is missing, enter it now.
- Use the Speed Annotation Tool to annotate multiple samples at once. Select annotations from the drop down boxes, and then select the samples in the text box, which the annotations should be applied to, and press Assign To Selected. Or press Assign To All.
- If you require more annotation fields use the Add Annotation Field drop down to select another annotation, then click Add to add the annotation field. If the annotation needed is not listed, select Other....'
- Once you are satisfied with your annotations, Click on Next Step (Sample Containers)
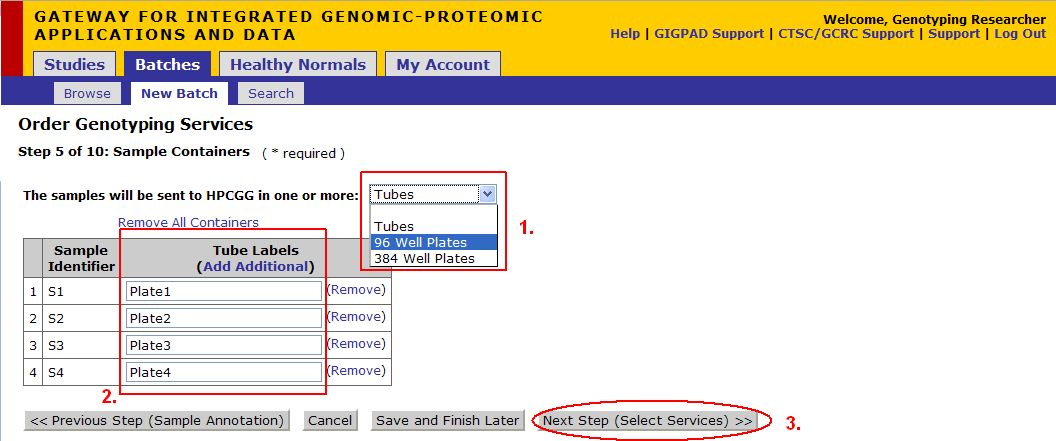
Step 5 of 10: Sample Containers
*Note: Sequonom orders must be submitted as Plates.
- Select the method in which the samples will be sent the Center, if you are submitting a Sequonom order your samples must be submitted as Plates.
- If you select plates as your method, you will be prompted to enter a name for the plate and click Add. If you would like to add multiple well plates, fill out the Add a Well Plate Field and click Add'.
- You can individually edit sample labels here
- Once you are satisfied with your container configuration click Next Step (Select Services)
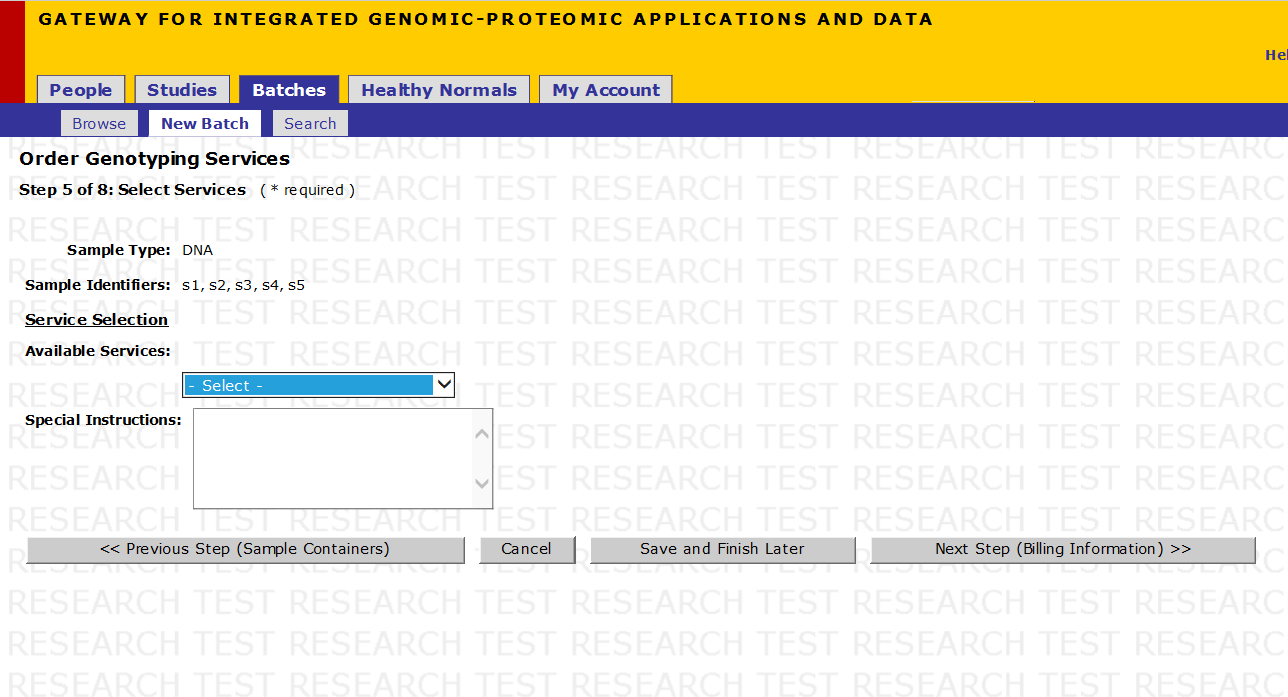
Step 6 of 10: Select Services
- Select Assay design and ordering from the Available Services if you require us to order oligos for your work, if you already have oligos stored at PCPGM then do not check this box
- For Genotyping Service, you will have been told during the assay design stage whether your designs are hME or iPLEX, please ensure that you select the appropriate genotyping chemistry from the drop down menu.
- Fill in the Special Instructions field if needed (up to 1000 characters)
- Click on Next Step (Billing Information)
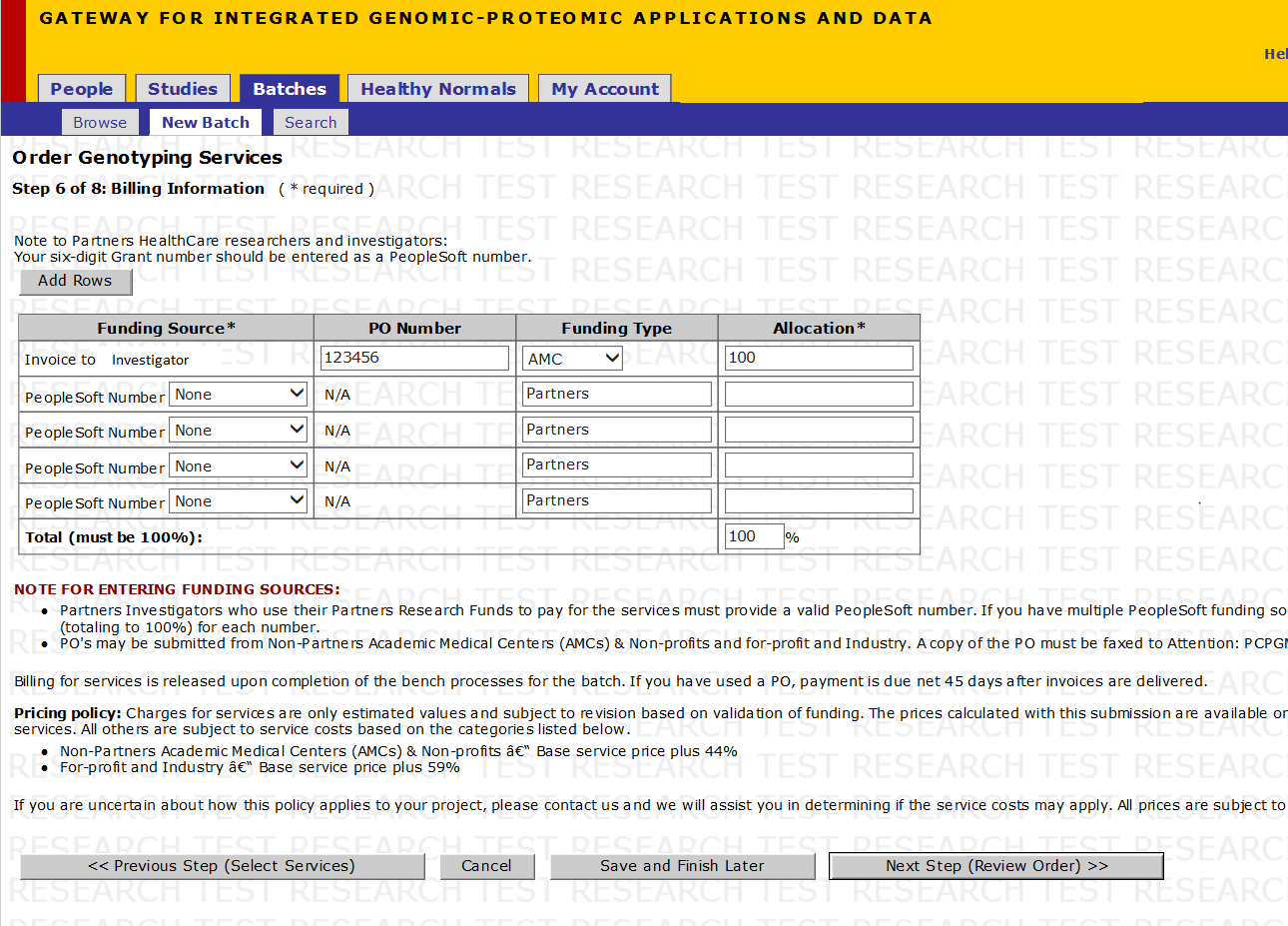
Step 7 of 10: Billing Information
Note:
- PO Number is associated with Invoice Funding Source only (not PeopleSoft)
- The Invoices are set up under the account of the PI associated with the Study as the PI’s Invoice Address. Please see the next page for details on how to set up an Invoice.
- You may enter as many Funding Sources as needed in any combination of PeopleSoft numbers or Invoices, as long as the Allocation equals 100%
- If you have an invoice set-up, enter the PO number and Allocation for that Invoice.
- If you would like to use a PeopleSoft Number(s) (For Partners Researchers Only)
- Select the PeopleSoft number from the drop down menu. If it is not present:
- To enter a new PeopleSoft number, select Add Number... from the drop down menu
- After all Funding Sources have been entered, the Allocation percentage in each row must total 100%.
- Click Next Step (SNP Selection)
Setting Up an Invoice Funding Source
If PO Number allocation exists, please fax copy of your PO document to
Fax: 617-768-8510
Attention: Alison Brown
Orders will not be approved until PO information is received by PCPGM. The PO information must be associated with the PI.
We will need the following information associated with the PO number.
- Physical Invoice Label
- Contact First Name
- Contact Last Name
- Address
- Phone Number
- Fax Number (Optional)
- Email Address
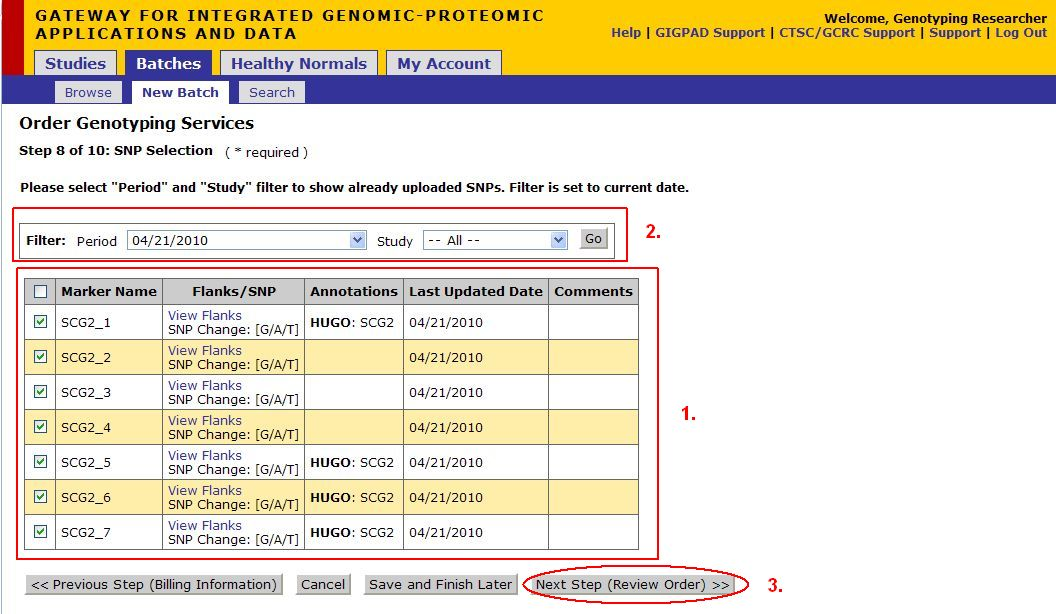
Step 8 of 10: SNP Selection
- The SNPs information uploaded during the SNP Submission step are shown here and are the SNPs that were uploaded during this Order Entry are automatically checked.
- If you are selecting SNPs that were previously uploaded. Use the Filter Buttons to select the Time Period and Study that the SNPs are associated with. Check all SNPs that you would like to use.
- When you have finished your selection, click on Next Step (Review Order)
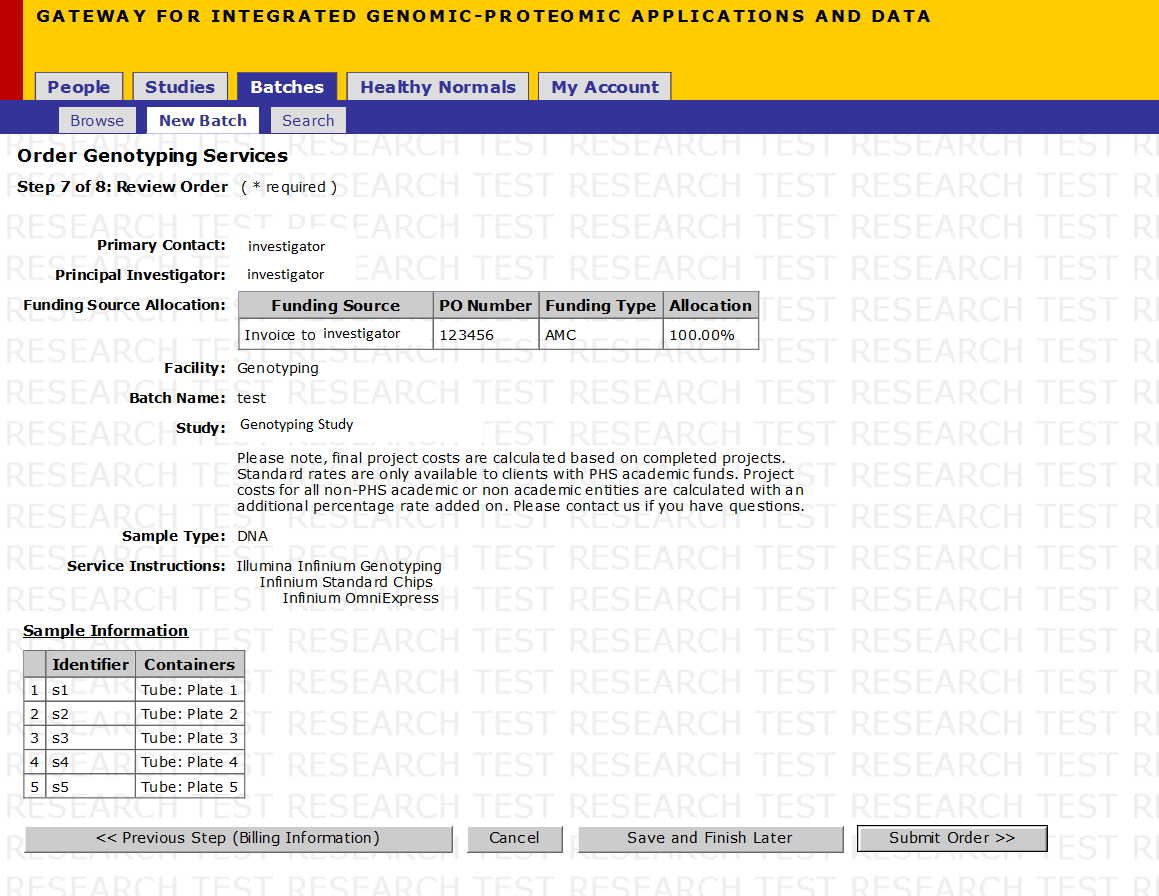
Step 9 of 10: Review Order
Review your order:
- Click on the Export link to export markers into an excel sheet
- If you are satisfied with your order click Submit Order
- If you need to edit your information click on Previous Step (Billing Information) to go to the previous step. Continue clicking on Previous Step until you reach the step you need to edit
- If you would like to finish the study later, click on Save and Finish Later to return to your order at a later time (Your order will be saved on the Batches->New Batch page)
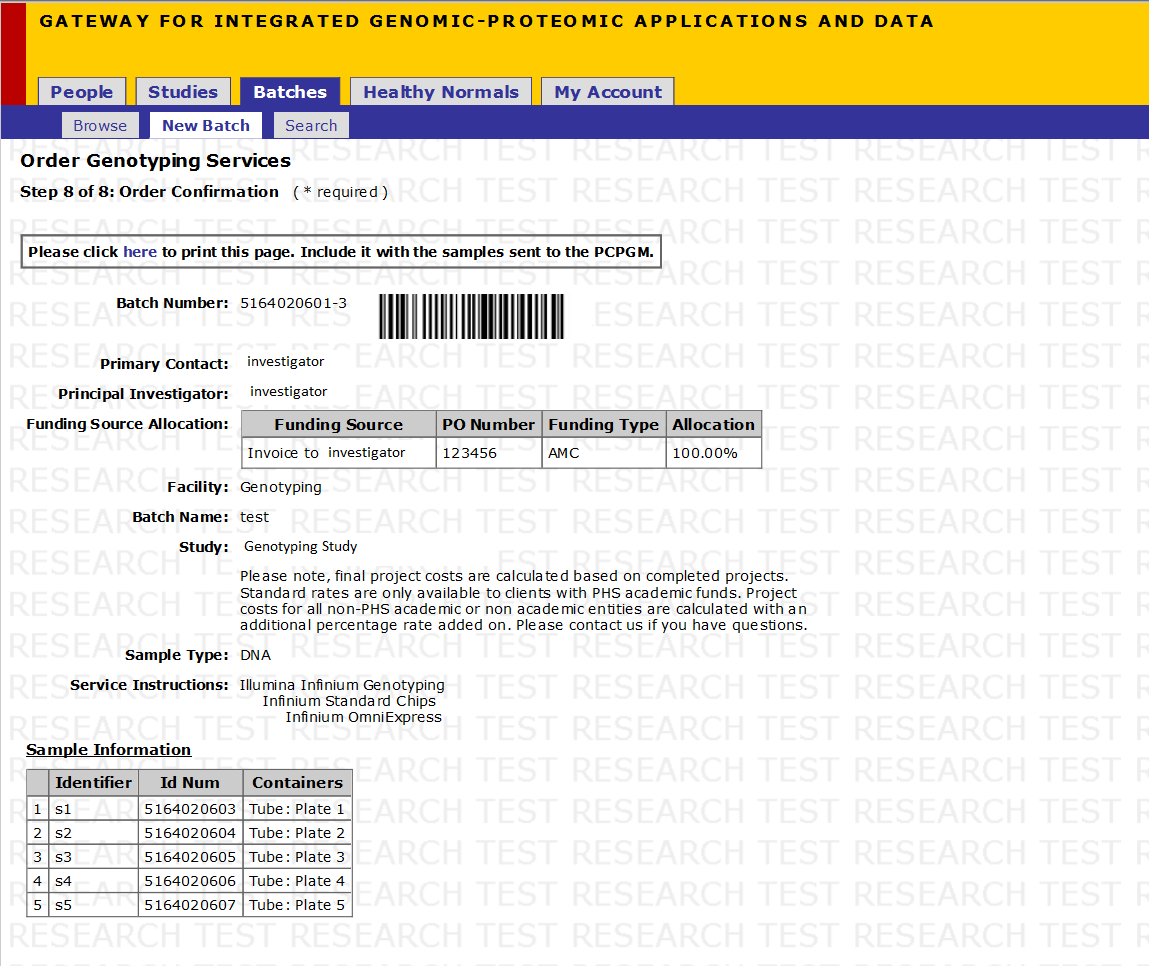
Step 10 of 10: Order Confirmation
- Click on the Export link to export markers into an excel sheet
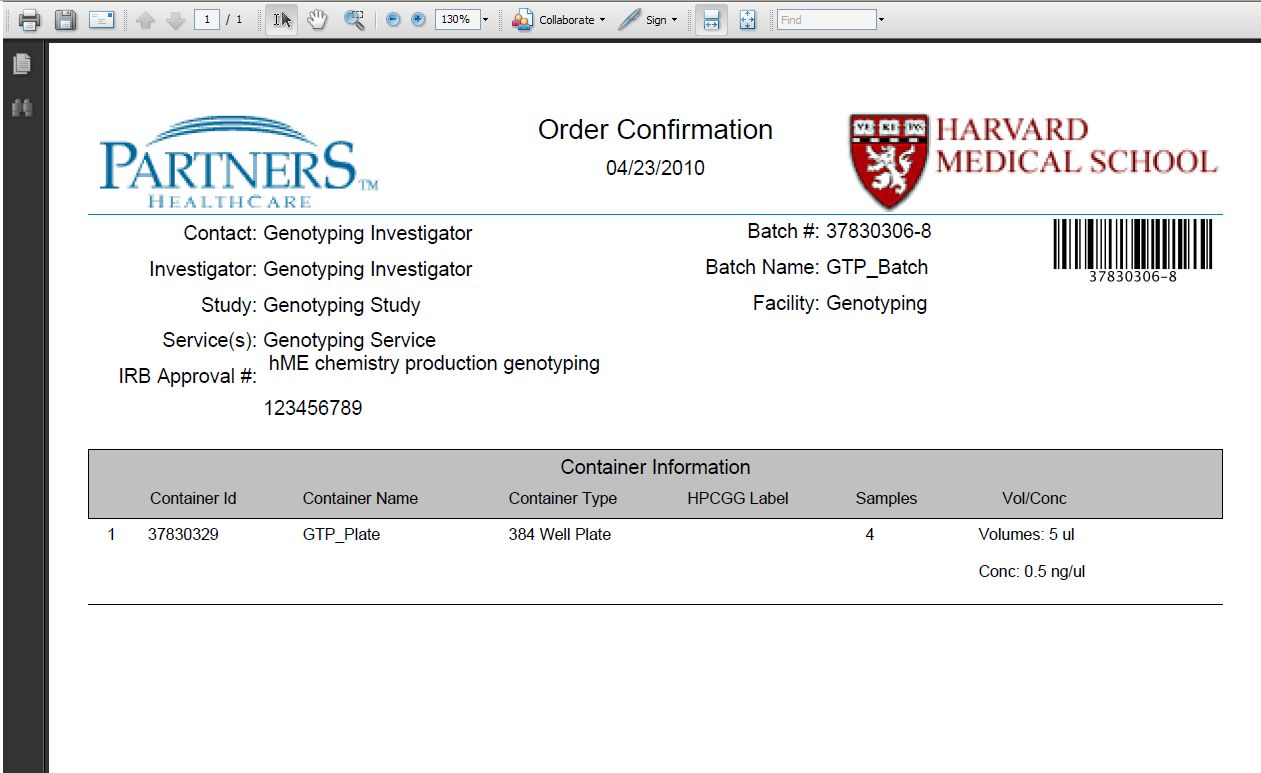
- Click on the here link to generate a printer-friendly PDF version of the order confirmation. Include this document with the samples that you send
Print the Order Confirmation Page
*Note: Please include this Order Confirmation page with the samples that are sent.